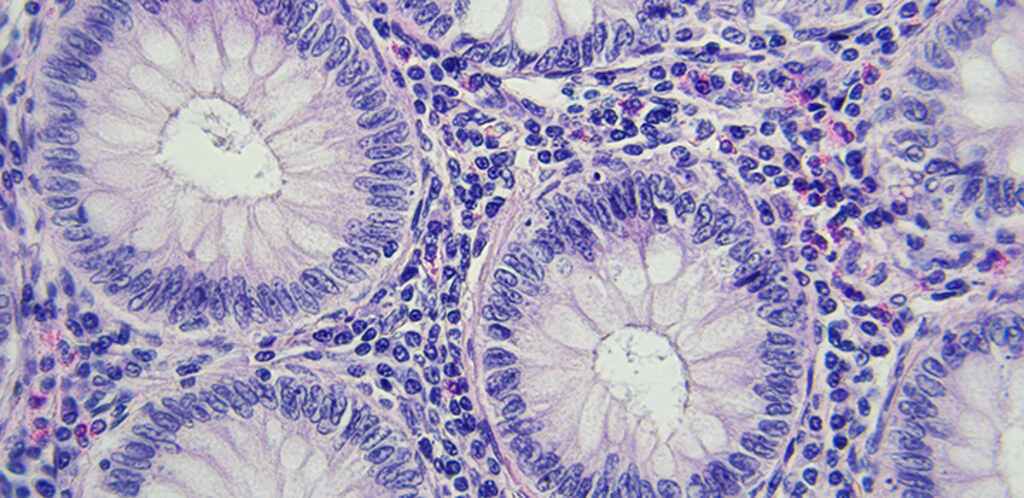
Research from the University of East Anglia reveals that colorectal cancer carries a unique microbial signature that distinguishes it from all other types of cancer. This finding contradicts earlier scientific beliefs that suggested every cancer type had its own microbial pattern.
Colorectal cancer affects the colon or rectum and is the fourth most common cancer in the UK. It’s also the second deadliest. This new finding could make a real difference in how doctors diagnose and treat this disease.
Scientists examined genetic information from 11,735 tumor samples across 22 cancer varieties. The team developed custom software to filter out human genetic material and isolate the microbial DNA present in each sample.
“This study changes how we think about the role of microbes in cancer,” said lead researcher Dr. Abraham Gihawi from UEA’s Norwich Medical School. “Our results show that only colorectal tumours possess distinctly identifiable microbial communities.”
What makes this discovery especially useful is that hospitals are already starting to use whole genome sequencing, which reads all of a person’s DNA. Now, doctors can look at the microbes in these same tests without extra cost.
More Posts
“We found that these microbial signatures were so specific that they could accurately distinguish colorectal tumours from other tumours,” Dr. Gihawi explained. The research suggests this approach could become a valuable addition to colorectal cancer diagnostics in the future.
The research also revealed benefits beyond just colorectal cancer. In oral cancers, the team found they could accurately detect HPV (human papillomavirus) compared to tests currently used. They also spotted rare but dangerous viruses like HTLV-1, which can hide in the body before causing cancer.
In sarcoma, a less common type of cancer, the researchers made an interesting discovery. “We found that certain types of bacteria were associated with poorer survival rates in some cases of sarcoma,” noted Dr. Gihawi. “This might lead to additional research and treatment options.”
The research uncovered that particular bacteria correlated with improved survival outcomes in specific sarcoma patients. This observation indicates that microbial profiles could potentially serve as indicators for treatment effectiveness in the future.
Professor Daniel Brewer from UEA emphasized, “This study highlights the growing clinical value of whole genome sequencing in identifying pathogenic organisms which may otherwise go undetected. By revealing these hidden infections and providing insight into cancer prognosis, genomic analysis is becoming an indispensable tool in precision medicine.”
The research involved many partners including the University of Leeds, the Quadram Institute, Oxford Nanopore Technologies, and others. It was funded by several organizations including the Big C Cancer Charity and Prostate Cancer UK. The findings were published on September 4, 2025, in the journal Science Translational Medicine.